Server Controls
Camera
In the server scene you have several interaction possibilities. In the case of multiple connected clients, the buttons on the bottom right labeled next and previous let you switch your current view between the clients. The view then shows what the selected client sees.
You can also change to a specific client’s view by clicking on the corresponding entry in the user panel on the left.
Loading and Saving
In the bottom right, there are two additional buttons Load and Save. The Load button opens a file dialog and lets you import all file formats supported by OpenBabel. The loaded molecule appears in front of the currently selected client. You can select a molecule in the active scene under the section AtomWorld and press the Save button to store the data in a file. Here, you have the option to pick a supported output format (by adding the corresponding file extension) and the data is automatically converted and saved in this format.
Creating Molecules
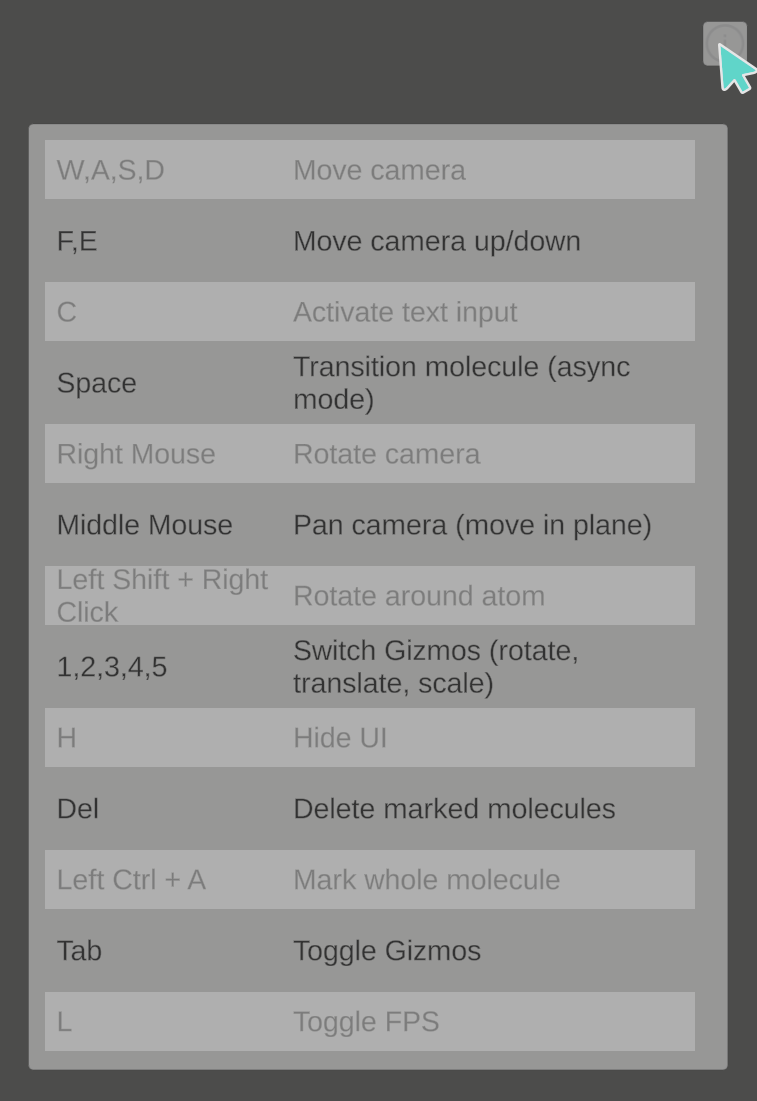
Pressing the C key on your keyboard activates a text input field. Entering any valid element symbol SMILES string and pressing the Enter key creates the specified molecule.
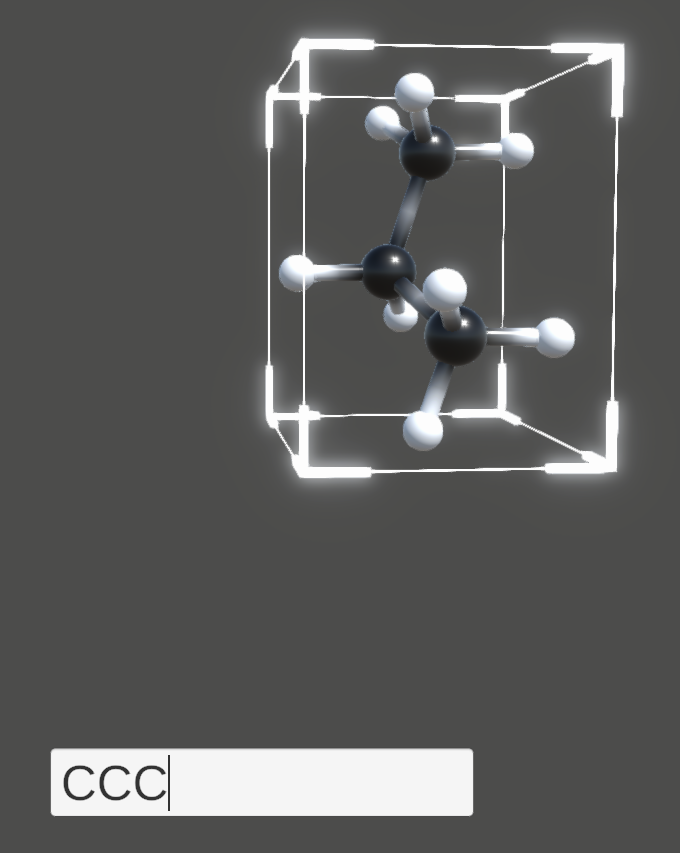
Pressing the Clear button at the bottom of the screen deletes all molecules in the scene.
Settings
A settings menu is activated by pressing the Settings button; it contains separate pages with different categories of settings. This includes settings for the force field, the UI, networking and more. Choosing your desired settings and pressing the Save button in the menu updates the settings both in the server scene and on any connected devices, if applicable.
Interacting with Molecules
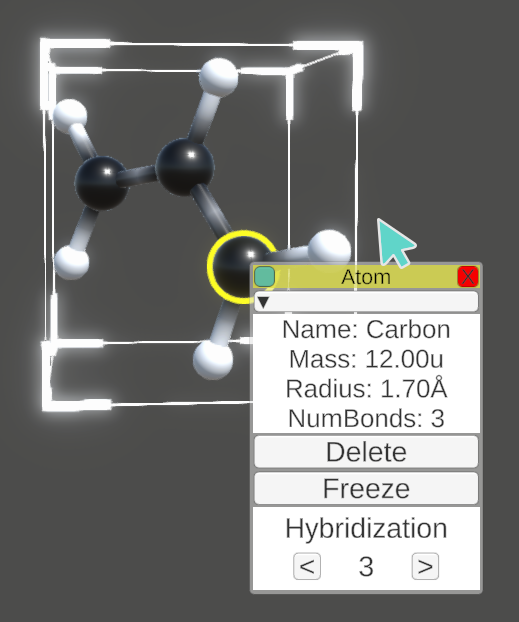
In the server scene, molecules can be interacted with using the mouse in a similar way to the AR environment. Clicking on an atom highlights it and displays a tooltip with information. Subsequent clicks on connected atoms display information about bonds, angles and torsion terms. Whole molecules can also be selected by clicking on a corner of their bounding box.
Tooltips can be dragged around the screen on the top bar or collapsed to save space.
Structure formulas
When selecting a whole molecule, the tooltip offers the additional option to generate a structure formula for the molecule using a Python library.
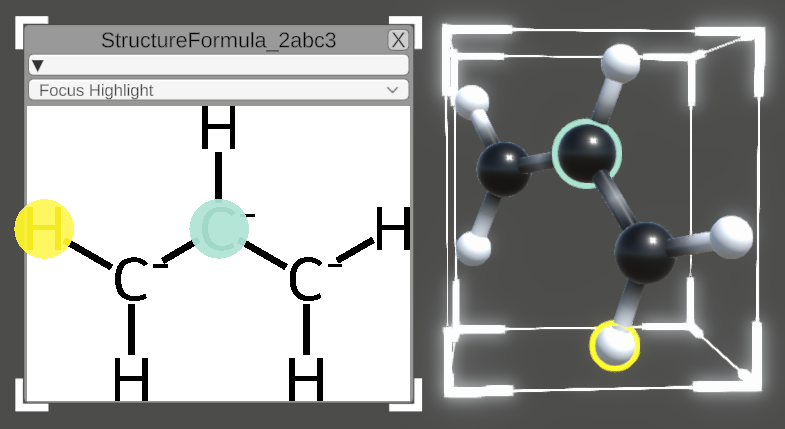
The structure formula window can be dragged (similar to the tooltip) or resized using the indicated corners. Atoms can be selected within the structure formula (turquoise) or within the molecule (default selection colors). The resulting highlighting is shown in both representations on corresponding structures.
Instead of displaying this highlighting, you may also choose to show a heatmap on the structure formula, which can for example be obtained by using eye-tracking data.
Transforming Molecules
Selecting a molecule creates a Gizmo consisting of three colored arrows in its center (this can be toggled on or off using the Tab key). Clicking and dragging one of the arrows moves the entire molecule along this line, while clicking the cube in the center allows to freely move the molecule. You can switch to similar Gizmos for rotating or scaling the molecule by pressing the number keys on the keyboard (1 for translation, 2 for rotation, 3 for scaling, 4, 5 and 6 for combinations).
Scaling a molecule non-uniformly (stretching it more along one axis than others) will cause unexpected behavior!
Other interactions
There are more shortcuts available to perform specific actions on the server. Hovering your mouse over the Info icon in the top right corner shows you a list of them.